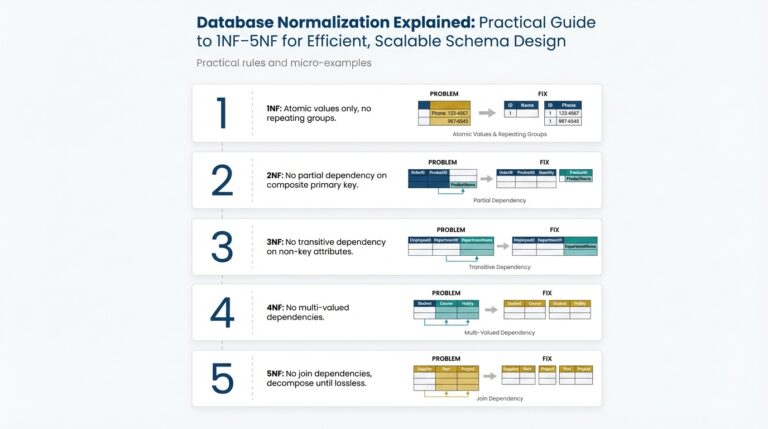
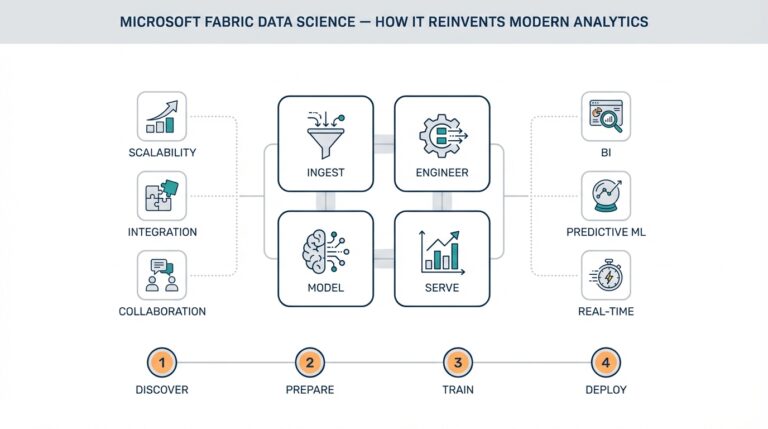
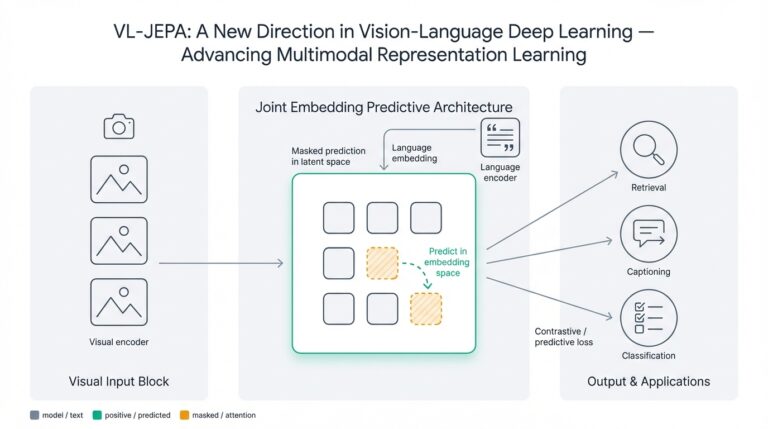
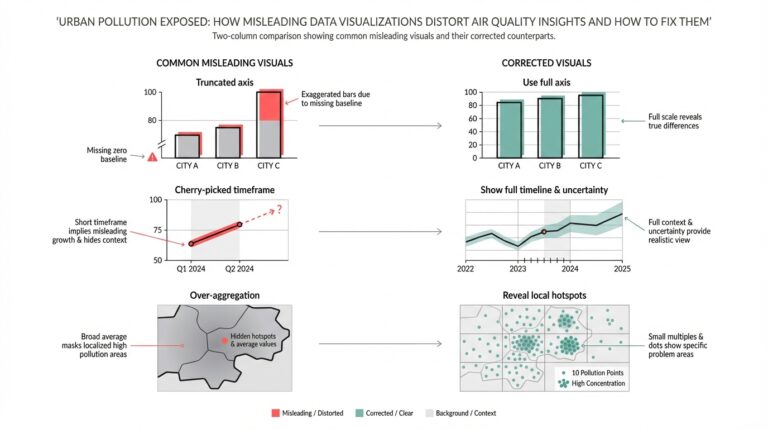
Understanding NLP and Its Role in Medicine
Natural Language Processing (NLP) is a field at the intersection of computer science, artificial intelligence, and linguistics that focuses on enabling machines to understand and interact with human language. In medicine, NLP plays a transformative role by helping decode the immense volume of unstructured text data generated daily in clinical notes, research articles, and patient records. This is particularly crucial because much of clinical documentation is recorded in the form of free-text, which can vary greatly in expression but still carry the same underlying meaning.
NLP algorithms analyze this data, extracting meaningful information to support numerous healthcare applications. For instance, automated systems can identify potential drug interactions, suggest diagnoses, or highlight relevant clinical trial opportunities based on patient records—all by interpreting language in medical documents. These capabilities support practitioners and researchers by significantly reducing the time spent on manual data review and by surfacing critical insights that may otherwise remain hidden.
One core challenge in applying NLP to medicine is the diversity of medical language. Medical professionals often use different terms, abbreviations, or even regional jargon to refer to the same condition or procedure. For example, “myocardial infarction” may appear in documentation as “heart attack,” “MI,” or simply “cardiac event.” Sophisticated NLP techniques—such as named entity recognition and context-aware language models—must be employed to bridge these linguistic differences and ensure accurate interpretation. The U.S. National Library of Medicine emphasizes the importance of standardized vocabulary in clinical practice for this very reason.
Moreover, the integration of NLP into healthcare systems is not just about technology; it’s about enhancing care delivery. By processing and piecing together diverse medical expressions, NLP tools enable clinicians to better aggregate patient histories, identify disease progression, and personalize treatment plans. For a closer look at how NLP is shaping real-world healthcare, HealthIT.gov offers insights into ongoing initiatives and case studies.
To summarize, NLP’s role in medicine is foundational to unlocking the potential of big data. It enables machines to understand nuanced clinical language, paving the way for improved decision-making, research, and ultimately, patient outcomes. For those interested in technical overviews, this academic review offers an in-depth exploration of NLP approaches and challenges in the medical domain.
Why Synonyms and Terminology Variations Matter in Healthcare Data
In the fast-evolving world of healthcare, data is both an asset and a challenge. One of the less obvious, yet critical, challenges lies in how medical information is recorded and interpreted—often using different words that mean the same thing. This issue of synonyms and terminology variations isn’t just a linguistic quirk; it’s a formidable barrier to effective data analysis, patient care, and even medical research.
Healthcare providers and researchers frequently encounter variations in how conditions, medications, and procedures are described. For example, “heart attack” and “myocardial infarction” refer to the same clinical event, but the choice of words can vary by specialist, institution, or even country. While these differences might not be problematic in conversation, they can create significant hurdles when aggregated in electronic health records (EHRs), analyzed for trends, or used to power machine learning models in natural language processing (NLP).
The Consequences of Inconsistent Medical Language
When clinical data is inconsistent, the accuracy of any system relying on that data is at risk. NLP algorithms, which are increasingly used to extract insights from vast troves of medical records, depend heavily on recognizing all synonyms and variations of a term. If the system fails to recognize that “dyspnea” and “shortness of breath” are identical, important connections can be lost—potentially impacting patient diagnosis and care pathways. Studies have shown that clinical NLP models struggle with terminology diversity due to the sheer volume of medical synonyms and acronyms.
Real-World Examples and Impact
Consider pharmacology, where a single drug may have multiple names: the chemical name, the generic name, and a host of brand names. “Acetaminophen” and “paracetamol” are chemically identical, but called different things in the US and the UK. If a medical records system can’t map these synonyms together, it could miss critical drug interactions, overprescribing, or adverse reactions—posing a real threat to patient safety. The same applies to procedures; “CABG” and “coronary artery bypass grafting” must be understood as one and the same procedure.
This fragmentation of medical language can also affect research outcomes. Meta-analyses, for example, depend on data from multiple sources. Terminology mismatches can lead to incomplete datasets, skewing results and potentially distorting clinical guidelines. The impact is so significant that the National Library of Medicine and the Unified Medical Language System (UMLS) have developed extensive resources and ontologies to facilitate consistent interpretation of medical language.
Practical Steps and Solutions
- Standardization: The adoption of standardized terminologies like ICD-10, MeSH, and SNOMED CT helps unify language across systems. These tools serve as reference points to map synonyms and ensure uniform data entry.
- Terminology Mapping: NLP tools are now designed to recognize, map, and harmonize synonyms using medical ontologies. For example, if an EHR contains “MI” (short for myocardial infarction), a sophisticated NLP model can map it back to its clinical synonym “heart attack,” ensuring accurate data aggregation.
- Continual Training: Medical language evolves rapidly, with new terms and abbreviations entering everyday use. Regularly training NLP systems with updated terminologies, clinical notes, and expert input is essential for maintaining accuracy.
Looking Ahead: The Human and AI Partnership
Succeeding in healthcare NLP isn’t just about algorithms; it’s about ongoing partnership between clinicians, informaticians, and data scientists. By consistently paying attention to synonyms and terminology, and investing in robust data infrastructure, we can ensure that critical medical insights are never lost in translation. For readers interested in the technical aspects, the UMLS Metathesaurus is a profound resource on this subject, providing structured mappings across nearly all medical vocabularies in use today.
Real-World Examples: When Different Words Mean the Same Thing
Consider a routine scenario in the emergency department: a doctor notes that a patient is experiencing “myocardial infarction,” while another refers to it as a “heart attack.” For human clinicians, these terms are immediate synonyms. However, for a natural language processing (NLP) system, especially one not finely tuned for medical jargon, this simple equivalence can become a major stumbling block. The challenge lies in accurately mapping different terminologies to the same clinical concept so that vital pieces of information are not overlooked.
Electronic Health Records: The Lexicon Dilemma
Within electronic health records (EHRs), the same condition can be documented in a multitude of ways. For instance, “hypertension,” “high blood pressure,” and even the shorthand “HTN” all refer to the same diagnosis. If an NLP tool isn’t trained to recognize these variations, critical data for research, billing, or patient care can be missed. Studies by the National Library of Medicine highlight the complexity and diversity of medical vocabularies and the necessity for advanced algorithms to decipher them.
Prescription Variations and Drug Names
Medications present another landscape of linguistic diversity: a drug may be documented by its brand name — like “Tylenol” — or its generic name, “acetaminophen.” Moreover, abbreviations such as “APAP” (for acetaminophen) further complicate extraction efforts. Failure to bridge these linguistic gaps can lead to duplicate data or, worse, potential clinical errors. An article by JAMA discusses how these name mismatches can result in adverse drug events when systems aren’t harmonized.
Diagnoses Described with Varying Specificity
Consider a scenario where one clinician documents “Type 2 diabetes mellitus,” while another simply writes “adult-onset diabetes.” Although both describe the same chronic disease, these nuances matter for NLP-driven analytics. To further complicate matters, variations can be driven by cultural or institutional preferences — e.g., “COPD” versus “chronic obstructive pulmonary disease.” Organizations like the Health Level Seven International (HL7) are working on standardized terminologies to address such issues, but the challenge persists in legacy data and free-text clinical notes.
Impacts on Research and Population Health
When NLP misses semantic equivalence, large-scale studies can yield inaccurate results. For example, if an algorithm only flags “congestive heart failure” but ignores “CHF” or “heart failure,” researchers might underestimate the prevalence of the condition. This was highlighted in a study that showed how misalignment in terms can distort outcomes, potentially affecting public health interventions and policy decisions.
Overall, these real-world examples reveal just how critical it is for NLP in medicine to handle linguistic diversity and synonymy. As healthcare data continues to grow in volume and complexity, developing robust systems that can interpret different words as the same meaning is not just an academic pursuit—it has tangible effects on patient safety, research quality, and healthcare delivery.
How Standardizing Medical Language Empowers Better Patient Care
One of the biggest obstacles in modern healthcare is the sheer diversity and complexity of medical language. Medical terminology isn’t just expansive – it’s nuanced, regional, and often specialty-specific. This can result in situations where two physicians describe the same condition or treatment using entirely different words. Such linguistic variation can create confusion, hinder data sharing, and ultimately impact patient outcomes. That’s where language standardization comes into play, bridging gaps across systems and professionals for more streamlined care.
The Case for Uniform Medical Language
When health professionals apply different terms for the same clinical concepts, crucial details can be lost in translation. For example, one doctor might chart “myocardial infarction,” while another writes “heart attack.” While both mean the same, automated systems and cross-institutional data sharing may not always recognize these as identical events unless there’s a common structure. This is why standardized vocabularies like UMLS (Unified Medical Language System) and SNOMED CT exist—to map out synonyms and ensure every term links back to a recognized medical concept.
Benefits for Clinicians and Patients
When language is standardized, healthcare records are easier to interpret, analyze, and exchange between institutions and even across borders. For example:
- Improved Decision Support: Electronic Health Record (EHR) systems can better alert clinicians to allergies, drug interactions, or care gaps when all terms are aligned. This leads to safer, faster interventions (source).
- Enhanced Research Opportunities: Aggregating data for studies and public health surveillance becomes easier, expediting breakthroughs or early identification of disease patterns. The CDC highlights just how crucial uniform records are for disease tracking.
- Better Patient Experience: Patients benefit from seamless care transitions. If their records are understood universally, repetitive tests or explanations can be avoided, leading to more satisfying experiences.
Real-World Challenges and Solutions
Standardizing language is not as simple as enforcing a single dictionary. Medical language evolves rapidly, and specialties, regions, and individual clinicians often develop preferred jargon. Successfully implementing standardized terminology often involves several steps:
- Mapping Local Terms: Start by identifying common synonyms and coding them to standard entries using resources such as the LOINC system for lab results.
- Training & Engagement: Clinicians need ongoing education about why standard vocabularies matter and how to use them efficiently without disrupting their workflow.
- Iterative Updates: Periodically review and expand standard lists to accommodate new treatments, discoveries, and shifts in language use. Organizations like HL7 International facilitate global collaboration in this continual process.
Looking Ahead: Technology as an Enabler
Natural Language Processing (NLP) is transforming how standardized language is put into practice. NLP tools can interpret free-text notes, recognize context, and translate variable expressions into standardized formats. For example, these systems can automatically equate “MI,” “heart attack,” and “myocardial infarction” so that all are recognized as the same diagnosis in a patient’s record. This process not only empowers accurate analytics and predictive modeling but also ensures continuity of care no matter where the patient goes.
Ultimately, embracing standardized medical language is not just about efficiency or data management—it’s about creating a healthcare system where every word leads to better, safer care. A truly patient-centered approach rests on clear, consistent communication, and language standards serve as its backbone.
Challenges in Training NLP Models for Medical Contexts
Building Natural Language Processing (NLP) models that truly understand language in medical contexts is far from straightforward. The complexity arises from several unique challenges that can derail even the most sophisticated AI systems if they aren’t addressed with care. Here’s an in-depth look at the major barriers encountered during the training of NLP models for medical applications.
Medical Terminology: A Moving Target
Medical language isn’t static. New terms appear with emerging diseases, drugs, and procedures, while older terminology may persist in clinical notes for decades. Training an NLP model requires grappling with this evolution. For example, the terminology for COVID-19 did not exist prior to 2019, and its rapid inclusion in academic literature posed a challenge to existing models. Standard abbreviations like “MI” could mean “myocardial infarction” or “mitral insufficiency,” depending on context.
To address these hurdles, models require regular retraining on recent datasets and exposure to a diverse range of sources, such as clinical notes, research articles, and patient queries. This approach ensures they stay current and capable of deciphering both legacy and newfound terms.
Synonymy and Polysemy: Same Symptoms, Different Sayings
A single concept may be expressed in a myriad of ways. For instance, “heart attack,” “myocardial infarction,” and even “MI” all refer to the same condition. Conversely, some words change meanings according to context—a phenomenon called polysemy. The word “discharge” could reference a medical release from a hospital or a physiological fluid.
Robust models must be trained on diverse datasets annotated by expert clinicians to capture these nuances. Building comprehensive ontologies, such as UMLS (Unified Medical Language System), has been vital for connecting synonyms and context-specific meanings. However, even these resources can’t cover every colloquial or newly-coined term, requiring ongoing human oversight and improvement.
Data Annotation: The Expertise Barrier
Unlike general language models, medical NLP models need training data annotated by professionals familiar with the intricacies of clinical language. Annotations specify where medical terms, symptoms, diagnoses, or treatments occur in text and what they refer to. Yet, annotator agreement can be surprisingly low, as studies have shown even experienced clinicians may interpret ambiguous phrases differently (JAMA Network Open).
Effective annotation frequently requires multi-stage processes: initial tagging, expert review, and consensus-building. These steps extend timelines and add expense but are vital for high-quality datasets able to train accurate models. Semi-automated tools are being developed to assist humans, but full automation still faces major challenges in this nuanced domain.
Diverse Document Types and Data Quality
The format of medical text spans concise prescriptions, rambling progress notes, imaging reports, and even patient emails. Each contains a distinct vocabulary and writing style—some heavily abbreviated, others verbose and narrative. Models trained on one type rarely generalize well to another, making universal comprehension tricky.
Moreover, clinical notes often include misspellings, dictation errors, and shorthand unique to individual practitioners or regions. Poor data quality means that pre-processing—spelling correction, expansion of acronyms, and validation—becomes an essential, labor-intensive step (Journal of Biomedical Informatics).
Privacy, Bias, and Fairness Considerations
Medical data is protected by stringent privacy regulations, such as HIPAA in the United States, making large-scale data collection and model training a delicate task. Anonymization tools are improving, but they are not infallible, and errors could lead to privacy breaches—a key ethical concern highlighted by The New England Journal of Medicine.
Bias in training data can also lead to inequitable model performance. For example, if a model is trained predominantly on English-language medical records from urban hospitals, it may not perform well for rural populations or non-English speakers. Careful diversification of datasets and continuous bias assessment are critical to ensuring fairness and accuracy across various demographic groups.
Ultimately, these multifaceted challenges reveal why developing high-performing NLP models for medical contexts requires not only advanced algorithms but also interdisciplinary collaboration, rich and evolving datasets, and a deep commitment to ethical, unbiased AI.
Strategies to Address Semantic Variation in Clinical Texts
Addressing semantic variation in clinical texts is a multifaceted endeavor that involves leveraging both linguistic insights and technological advances. The challenge arises as different clinicians may use diverse terms or phrases to describe the same medical condition, treatment, or finding, complicating automated understanding by Natural Language Processing (NLP) systems. Here are several key strategies employed to tackle this hidden barrier in medical NLP:
Building and Expanding Medical Ontologies
One foundational approach is the creation and refinement of medical ontologies—structured frameworks that define the relationships between medical concepts. Resources like the Unified Medical Language System (UMLS) and SNOMED CT aggregate synonymous terms and map them to standardized codes. Implementing these in NLP workflows allows for normalization of terms—”heart attack,” “myocardial infarction,” and “cardiac infarction” are recognized as the same underlying concept. Regular updates and community contributions expand these ontologies to stay current with medical advances.
Leveraging Synonym Expansion and Entity Linking
Synonym expansion involves automatically replacing or supplementing words with their synonyms using curated lists or ontologies. This is often combined with entity linking, where clinical terms within texts are connected to their unique identifiers in medical databases. For example, mapping the phrase “HTN” to “hypertension” and linking it to a known clinical entity. These techniques ensure that semantic variation does not obscure the clinical meaning for downstream analysis, such as cohort identification or automated coding.
Context-Aware Language Models
Recent breakthroughs in NLP have come from context-aware language models such as BioBERT and SciBERT, which are specifically trained on large corpora of biomedical literature. These models excel in capturing nuanced meanings based on surrounding context, enabling them to discern subtle differences in usage and adapt to the variability in clinician language. For example, the word “discharge” can mean either fluid excretion or patient release—contextual models can determine the intended meaning based on sentence structure and neighboring terms.
Active Learning and Human-in-the-Loop Approaches
Given the complexity of clinical language and the importance of accuracy in healthcare, some systems employ active learning, where the NLP model highlights ambiguous or novel phrases. These instances are then reviewed and annotated by domain experts. Incorporating human feedback helps the model adapt to evolving terminology and rare synonym usage. For instance, an unfamiliar abbreviation detected in physician notes can be flagged, clarified by a clinician, and integrated into the system for future recognition—a process described in studies such as those highlighted by the National Center for Biotechnology Information (NCBI).
Domain-Specific Preprocessing Pipelines
Effective preprocessing can dramatically improve downstream NLP performance. Techniques may include abbreviation expansion, correcting misspellings, and segmenting compound medical terms. For instance, tools like NLTK and domain-tuned modules can parse unstructured notes and prepare them for semantic analysis. Carefully designed pipelines that account for domain-specific quirks—such as how “SOB” refers to “shortness of breath” rather than its colloquial meaning—help reduce errors due to semantic drift.
By combining these strategies, clinical NLP systems can more reliably interpret the wealth of information contained in electronic health records, research papers, and other healthcare documentation. These approaches are essential to ensure that the true meaning behind medical texts is understood, regardless of the words chosen by individual practitioners.