Why df.equals() fails
If you’ve ever written a quick sanity check like df1.equals(df2) and then been surprised by a False, you’re not alone — df.equals() is a strict, label-and-type-sensitive comparison and often fails for reasons that matter less in real-world validation. In the first 100 words we want to emphasize that df.equals(), Pandas, and PySpark behave very differently when you ask them to prove two tables are “the same.” Building on what we covered earlier about schema and row-order assumptions, this section explains the common mismatch patterns that make df.equals() unreliable for robust Dataframe Comparison in pipelines and tests.
At its core, Pandas’ df.equals() does an elementwise equality check between two DataFrame objects and requires matching index labels, column labels, shapes, and element equality — NaNs in the same locations are treated as equal. For straightforward debugging this behavior is useful, but it’s strict: differing column order, a different index, or mismatched dtypes will flip a True to False even when your data content is logically the same. For example:
# pandas example
# df1 has int64 id, df2 has float64 id but same numeric values
df1.equals(df2) # often False because dtypes differ
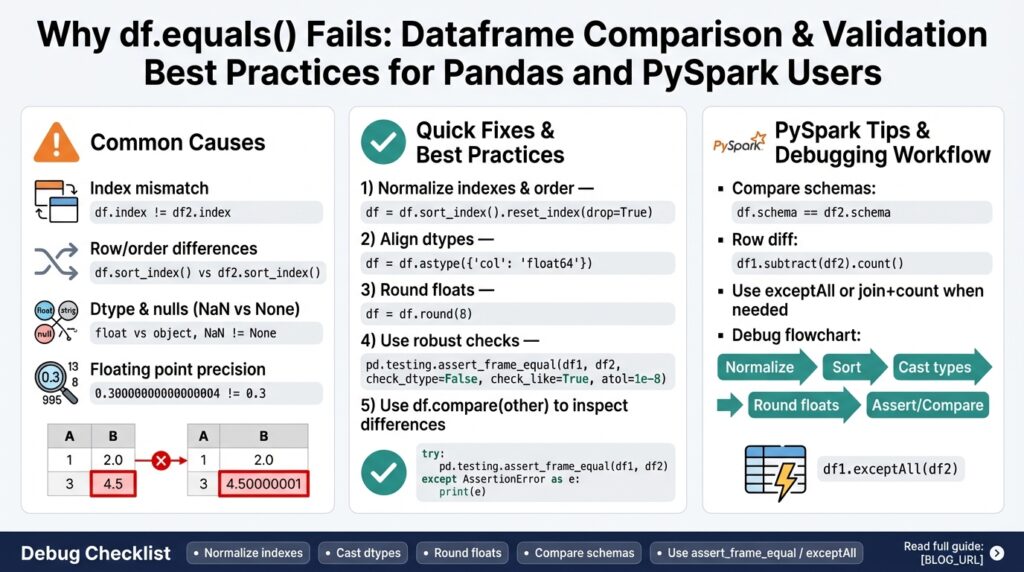
Several concrete failure modes recur in practice and explain most surprise False results. First, index and column ordering matters: two DataFrames with identical rows but different sort order or column order will not be equal. Second, dtype mismatches (int vs float, categorical vs object, timezone-aware vs naive datetimes) break equality even when values are numerically identical. Third, floating-point rounding and precision differences cause elementwise comparisons to fail; small arithmetic or serialization differences are enough to flip equality. Finally, presence of duplicate column names, mixed None/NaN representations, or hidden metadata can produce unexpected outcomes during Dataframe Comparison.
Floating-point and null semantics deserve special attention because they’re common in ETL and analytics workloads. df.equals() requires bitwise-equal floating values — it won’t treat 0.30000000000000004 and 0.3 as equal — so you should instead canonicalize numeric types and use tolerance-aware comparisons (numpy.isclose or pandas.testing.assert_frame_equal with rtol/atol) when small differences are acceptable. Likewise, df.equals() treats NaN==NaN as True, which can hide differences when one side uses None or a sentinel; normalize missing values explicitly before comparing. For example, prefer assertions like pandas.testing.assert_frame_equal(df1.sort_index(axis=1), df2.sort_index(axis=1), check_dtype=False, rtol=1e-5) for robust testing.
When you move from Pandas to distributed engines, the mismatch gets worse: PySpark DataFrame semantics aren’t the same as Pandas, and df.equals() is not a reliable distributed equality check. How do you compare large Spark DataFrames reliably? In Spark you must normalize schema, sort deterministically (or hash rows), and use set-difference operations such as exceptAll or left‑anti joins to find mismatches; collecting and comparing Python-side objects is only viable for small datasets because collect() can explode memory. In practice, compute stable row-level checksums or use repartition+sort keys then compare counts of diffs to validate equality at scale.
Given these pitfalls, adopt a reproducible comparison recipe: normalize column order and dtypes first, canonicalize nulls and datetimes, use tolerance-aware numeric comparisons, and prefer framework-specific techniques (pandas.testing utilities for Pandas; exceptAll/row-hash + counts for PySpark). Where performance or determinism matters, add multi-stage checks: compare schemas, row counts, hash summaries, and finally sampled or full row diffs. Taking this concept further, the next section will show concrete code patterns and helper functions you can drop into tests and ETL jobs to make Dataframe Comparison deterministic, explainable, and performant.
Common mismatch causes
You run df.equals() and get False — even though a quick visual inspection suggests the tables are identical. Building on the foundation we established earlier, start by asking: what causes df.equals() to return False even when your data looks the same? In practice, most surprises come from structural or representation differences rather than the underlying business values. Front-load your troubleshooting with schema and representation checks because DataFrame Comparison tools like df.equals() are strict about labels, dtypes, and hidden metadata.
One frequent culprit is ordering and duplicate labels. Two DataFrames with the same rows but different column order, index order, or duplicate column names will fail elementwise equality. For instance, swapping columns or sorting rows differently will flip df.equals() to False even when values match; similarly, duplicate column names can align wrong values during comparison. To diagnose this, verify sorted column lists and check DataFrame.columns. If you see duplicates, decide whether to rename, aggregate, or explicitly align by position before comparing.
Type and representation mismatches are another common root cause. Numeric types (int64 vs float64), categorical codes vs object strings, and Decimal precision/scale differences all break bitwise equality. For example, a column cast to float during CSV read or Parquet write will not be equal to the original integer column even if every value prints the same. In Pandas, inspect dtypes with df.dtypes; in PySpark, inspect the schema and nullable flags. Type normalisation—explicit casts or using nullable integer types in Pandas—often resolves these mismatches before you run a strict comparison.
Floating-point precision and small serialization differences create hard-to-see mismatches next. Floating values that differ by rounding (0.30000000000000004 vs 0.3) are not equal to df.equals(), since it performs elementwise bitwise checks. When you expect minor numeric drift from computations or serialization, adopt tolerance-aware checks (for example, compare hashes of rounded values or use isclose/assert_frame_equal with rtol/atol). This prevents noisy False results from harmless floating-point noise while still catching meaningful numeric divergence.
Null semantics and missing-value representation cause subtle false negatives. Pandas treats NaN==NaN as True in df.equals(), but None, NaT, or framework-specific nullable types (Pandas’ Int64 vs numpy int64, Spark’s nullable fields) can appear different. Additionally, missing keys in nested JSON or differences between an explicit null and an absent column in Spark schema evolution will break equality. Normalize nulls—use df.fillna or explicit casts to framework-nullable types—and ensure both sides use the same sentinel for missing values before you compare.
Distributed-engine peculiarities amplify these issues in PySpark. Row order is non-deterministic without a sort key, partitioning can hide duplicates or cause different result ordering, and Parquet/ORC schema evolution may promote types (e.g., int -> long). Nested structs and arrays can differ in field order or metadata even when semantic content is the same. For large datasets, avoid collect()-based comparisons; instead compute stable row-level checksums (hash concatenation of normalized columns), compare schema+counts, and then run set-difference (exceptAll or left-anti joins) to locate true mismatches.
Detecting the root cause efficiently requires a multi-stage approach: compare schemas and column orders first, then row counts, then column-wise dtype diffs, then lightweight checksums, and finally sampled or full row diffs. When you follow this sequence you turn a single False from df.equals() into actionable evidence about ordering, dtype, precision, nulls, or distribution issues. Taking these diagnostics into your tests and ETL pipelines makes DataFrame Comparison repeatable and explainable; next we’ll translate these checks into concrete code patterns and helper functions you can drop into your test suite or jobs.
Index, order, and dtypes
When df.equals() surprises you with False, the most common culprits are index alignment, row/column order, and dtype mismatches — these three often masquerade as content differences during DataFrame Comparison. Building on the schema-and-order diagnostics we described earlier, start by treating labels and types as first-class citizens: df.equals() requires identical index labels, identical column labels in the same order, and bitwise-equal element types. How do you know when index, order, or dtype differences are behind a spurious df.equals() == False? If the values look the same on a quick sample but equality fails, assume one of these three structural issues until proven otherwise.
Index mismatches are deceptively common in ETL and tests. If one side uses a natural key as the index and the other uses a 0..n range, elementwise comparison will misalign rows even when every row’s payload matches; reset_index(drop=True) or using reset_index on both DataFrames before comparing forces positional alignment. For stable comparisons we often canonicalize index and sort rows deterministically: df1 = df1.reset_index(drop=True).sort_values(key_cols).reset_index(drop=True); df2 = df2.reset_index(drop=True).sort_values(key_cols).reset_index(drop=True). This removes label-based false negatives and makes downstream hash or elementwise checks meaningful.
Column order matters for df.equals() and for many hashing strategies, so normalize column layout before comparing. Sorting columns alphabetically or by a canonical schema list ensures the same visual and programmatic alignment: df = df[canonical_cols]; if duplicate column names exist, explicitly disambiguate them because positional alignment may silently compare the wrong columns. For wide tables where reordering is expensive, compare column-level checksums (e.g., df[col].astype(str).agg(‘|’.join) or hash functions) after casting to a canonical dtype to detect order-agnostic differences quickly.
Dtype mismatches are another frequent root cause and require careful decisions about canonicalization versus tolerance. Integer-to-float promotion, categorical versus object, and timezone-aware versus naive datetimes will flip df.equals() to False even when printed values match; explicitly cast both sides to a chosen dtype family before the final comparison (for example, df[col] = df[col].astype(“Int64”) or df[col] = df[col].astype(“float64”)). When numeric drift is acceptable, use tolerance-aware comparisons (numpy.isclose or pandas.testing.assert_frame_equal with rtol/atol) rather than forcing identical dtypes, because forcing floats into ints can hide legitimate NaN semantics.
When you work with PySpark, these structural issues scale and change shape: order is non-deterministic without an ORDER BY, partitioning affects row distribution, and schema evolution can promote types across writes and reads. In Spark, normalize schemas with explicit casts, enforce a deterministic sort key with orderBy, or compute stable row hashes to compare sets instead of relying on order. For example, compute a row checksum: df = df.selectExpr(“md5(concat_ws(‘|’, cast(col1 as string), cast(col2 as string))) as row_hash”), then compare counts of row_hash differences with exceptAll or join-based anti-diff to find true mismatches without collect().
Taking these normalization steps turns a single False from df.equals() into actionable evidence. First align index and sort rows, then normalize column order and names, then reconcile dtypes or apply tolerance-aware comparison for numerics and datetimes, and finally use checksums or framework-specific set-diff operations for large datasets. Taking this concept further, in the next section we’ll translate these rules into reusable comparison helpers and small, test-friendly functions you can drop into Pandas and PySpark pipelines to make DataFrame Comparison deterministic, explainable, and fast.
NaN and float tolerance
Floating-point drift and inconsistent null representations are the two silent reasons df.equals() flips from True to False in real workloads, so we need a practical, repeatable way to reconcile them. How do you reconcile NaN differences and tiny floating-point drift when df.equals() returns False? Building on the normalization and dtype lessons above, start by treating missing-value semantics and numeric tolerance as explicit parts of your comparison contract rather than incidental artifacts. In practice that means normalizing null sentinels, choosing a numeric tolerance strategy, and making those choices visible in tests and ETL assertions so failures point to business-significant divergence instead of incidental noise.
Null semantics differ across frameworks and formats, and that mismatch often hides real defects. In Pandas a column may contain numpy.nan, Python None, or a nullable integer type (“Int64”) where NA is intentional; in PySpark floats can contain NaN while other types use null; downstream reads from CSV/Parquet can promote or materialize different sentinels. Normalize both sides before comparing: coerce nullable integer columns to Pandas’ nullable dtypes (df[col] = df[col].astype("Int64")) or explicitly fill/flag missing values (df = df.fillna({'col': None}) or df = df.replace({None: np.nan})) depending on which semantics you must preserve. In Spark, use df.na.fill() for simple fills or coalesce() and explicit cast() to canonicalize fields before any hashing or set-diff operation.
Floating-point differences arise from arithmetic, serialization, and storage precision; treat them as expected but bounded. Use tolerance-aware, elementwise checks instead of bitwise equality: numpy.isclose for array-level checks and pandas.testing.assert_frame_equal with rtol/atol for DataFrame assertions give you control over acceptable drift. For example, in tests you might call pandas.testing.assert_frame_equal(df1.sort_index(axis=1), df2.sort_index(axis=1), check_dtype=False, check_like=True, rtol=1e-5, atol=1e-8) to ignore column order and tolerate tiny numeric noise. When rounding is an acceptable canonicalization strategy, apply df.round(n) for the required number of decimals before hashing or comparison to make differences explainable and repeatable.
For pipeline validation and large datasets, combine normalization with lightweight, deterministic summarization instead of full collect-based checks. Normalize nulls and cast floats to your canonical types, then compute stable row-level checksums using only normalized string casts and controlled rounding: from pyspark.sql.functions import md5, concat_ws, col; checksum_df = df.select(md5(concat_ws('|', *[col(c).cast('string') for c in cols])).alias('row_hash')) — this pattern gives you order-agnostic, scale-friendly comparison keys. Compare checksums with exceptAll or left-anti joins in Spark to find true mismatches; in Pandas, compare hashed rows or use merge with indicator to isolate differing records. This two-stage approach (normalize, then summarize/compare) prevents tiny float noise or differing NA representations from producing noisy test failures while still surfacing substantive data drift.
Choosing the numeric tolerance and null strategy is a product decision as much as a technical one, so codify it where tests and ETL jobs can read it. Ask yourself: do business rules require exact integer semantics, or can you accept a relative error (rtol) of 1e-6? When you need tighter guarantees, lower atol/rtol; when downstream aggregation amplifies noise, prefer rounding before comparison. Taking these choices out of ad-hoc scripts and into small, reusable helper functions makes DataFrame Comparison deterministic, auditable, and easier to debug — next we’ll translate these patterns into compact helpers and test fixtures you can drop into Pandas and PySpark pipelines.
Pandas vs PySpark comparisons
Building on this foundation, the first 100 words must make one thing clear: df.equals() lives in a different universe than large-scale, distributed comparison tools. In Pandas, df.equals() performs elementwise, label-sensitive equality and treats NaNs specially; in distributed engines like PySpark you instead reason in sets, partitions, and schema evolution. For robust DataFrame Comparison you need to pick strategies that respect each framework’s primitives rather than forcing one tool’s semantics onto the other.
At a high level, Pandas comparisons are object-local and index-aware, while Spark comparisons are set-oriented and partitioned. In Pandas you can rely on deterministic in-memory operations such as pandas.testing.assert_frame_equal, numpy.isclose, or explicit casts to reconcile dtype and floating-point issues. In PySpark, row order is non-deterministic without orderBy, and schema promotion (int→long, nullability changes) is common across reads and writes; therefore you use exceptAll, left-anti joins, or stable row-level hashes to detect differences at scale.
How do you choose the right comparison strategy for a given workload? If both DataFrames fit comfortably in memory, prefer Pandas-native, tolerance-aware checks: normalize columns, enforce canonical dtypes, sort deterministically, and then call pandas.testing.assert_frame_equal(..., check_dtype=False, rtol=..., atol=...). For datasets that live in Spark, compute a deterministic row checksum (for example md5(concat_ws('|', *cols)) after casting and rounding) and compare those checksums with exceptAll or an anti-join rather than pulling all rows to the driver. This keeps your validation memory-safe and order-agnostic.
Consider a pragmatic cross-framework scenario: an ETL job writes Parquet files from Spark and your unit tests read those files into Pandas for assertion. If you rely on df.equals() in Pandas without schema normalization you’ll see failures due to promoted types, timezone differences, or different null sentinels. Instead, we normalize on the Spark side (explicit cast()s and orderBy on canonical keys), export with predictable types, and then in Pandas coerce dtypes and apply df.round() or numpy.isclose where numeric drift is acceptable. These steps dramatically reduce spurious mismatches and make any remaining diffs actionable.
Performance and scalability shape the comparison choices you make. Avoid collect() for large Spark tables; collect() can blow memory and loses Spark’s distributed correctness guarantees. Use lightweight invariants first: compare schemas, counts, column-level min/max, and column histograms or hashed samples before attempting full row diffs. When full verification is required, compute and compare stable row hashes with repartition() on your sort key to amortize shuffle cost and then use exceptAll to enumerate true differences without pulling raw payloads to the driver.
Testing and CI integration benefit when you codify framework-specific comparison recipes as small helpers. In unit tests expose parameters for rtol/atol, canonical dtype maps, and permitted schema promotions so that test failures point to business-relevant changes rather than incidental representation drift. Log example mismatched rows or row-hash counts in a compact artifact that CI can surface; this gives developers immediate clues about whether a failure is structural (schema/order) or numeric (precision/tolerance).
Taking this concept further, we treat comparison as a multi-stage pipeline: schema alignment, dtype normalization, lightweight summary checks, and then deterministic, framework-native diffs. By aligning Pandas and Spark semantics where possible—explicit casts, null canonicalization, controlled rounding, and hash-based set-diffs—you convert a noisy df.equals() False into precise, explainable evidence. In the next section we’ll translate these rules into reusable helper functions and short code patterns you can drop into tests and ETL jobs to make DataFrame Comparison deterministic and auditable.
Robust comparison strategies
df.equals() and naïve elementwise checks are useful for quick sanity tests, but they fail fast in real pipelines—different column order, dtype promotion, tiny floating drift, or index misalignment will flip a True to False long before you’ve isolated a business-impacting divergence. How do you build a comparison that tolerates incidental noise but still flags meaningful data drift? Start with a reproducible, multi-stage strategy for DataFrame Comparison that treats schema, counts, summaries, and row-level identity as separate, verifiable invariants.
Begin by validating schema and cardinality because structural mismatches are the cheapest to detect and explain. First assert the canonical column set and nullable/nullable differences; then compare row counts and primary-key uniqueness. If either schema or counts differ, that’s a structural failure you should fix before deeper content checks. This approach reduces noisy downstream work and gives you an early, actionable signal about schema evolution, ingestion bugs, or accidental truncation.
Next, compute lightweight column-level summaries to catch category- or distribution-level drift without a full row diff. Compare min/max, distinct counts, null counts, and simple histograms for numeric columns; these invariants surface missing partitions, unexpected null inflation, and gross aggregation errors. When you need numeric tolerance, front-load it here by comparing rounded aggregates or percentiles (for example compare rounded means/medians using a defined tolerance) so you don’t escalate every tiny floating difference unnecessarily.
For deterministic row-level checks at scale, canonicalize representations and then use stable row hashes instead of elementwise comparisons. In Pandas normalize by resetting indexes, ordering columns consistently, casting to canonical dtypes, and then compute a row hash: df['row_hash'] = df[canonical_cols].astype(str).agg('|'.join, axis=1).apply(lambda s: hashlib.md5(s.encode()).hexdigest()) — compare sets of hashes or merge with an indicator to isolate mismatches. In-memory tests can still use pandas.testing.assert_frame_equal(..., check_dtype=False, check_like=True, rtol=1e-5, atol=1e-8) after the same normalization steps when you want richer assertion failures rather than just counts.
When you work with PySpark, apply the same normalization but use distributed primitives: cast and round columns to canonical types, repartition on a deterministic key, and compute a stable checksum with SQL expressions (for example md5(concat_ws('|', col1.cast('string'), col2.cast('string'), ...))). Compare those checksums with exceptAll or a left-anti join to enumerate true mismatched rows without pulling the whole dataset to the driver. Avoid collect() for large tables; instead compare checksum counts and sample mismatches for debugging. This set-oriented approach respects Spark’s partitioned execution model while keeping comparisons memory-safe and explainable.
Float tolerance and null semantics must be codified as policy, not ad-hoc choices. Decide when to use elementwise tolerances (numpy.isclose/rtol+atol), when to round before hashing, and when to treat None vs NaN as equivalent; encode those decisions in helper functions so tests and ETL jobs speak the same language. Finally, operationalize these stages into small utilities that your CI can run: schema check, count check, summary diffs, hash set-diff, then sampled payload diff. Taking this multi-stage, canonicalization-first approach turns a noisy df.equals() False into precise, actionable evidence you can automate and debug; next we’ll translate these patterns into compact, reusable helpers you can drop into tests and ETL jobs.